Repeated protein domains
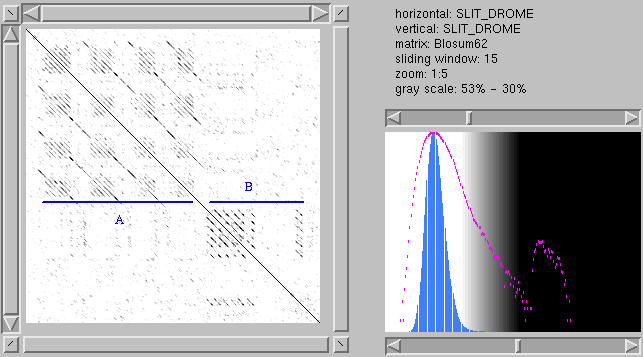
This is a plot of Drosophila melanogaster
SLIT protein against itself. It has several
repeated domains. In the N-terminal part (A), we
see four repeated regions, which are themselves
made up of smaller repeated units (in this case,
leucine-rich repeats). Then, there is another
domain that's repeated at least six times in a
tight cluster (B), with one additional occurrence
near the C-terminus. This one is an EGF.
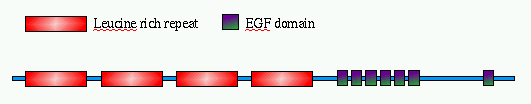
The figure below shows the arrangement of the
domains along the protein sequence, as described
in the Swiss-Prot entry.
SLIT_DROME (P24014):
MAAPSRTTLMPPPFRLQLRLLILPILLLLRHDAVHAEPYSGGFGSSAVSSGGLGSVGIHIPGGGVGVITEARCPRVCSCT
GLNVDCSHRGLTSVPRKISADVERLELQGNNLTVIYETDFQRLTKLRMLQLTDNQIHTIERNSFQDLVSLERLDISNNVI
TTVGRRVFKGAQSLRSLQLDNNQITCLDEHAFKGLVELEILTLNNNNLTSLPHNIFGGLGRLRALRLSDNPFACDCHLSW
LSRFLRSATRLAPYTRCQSPSQLKGQNVADLHDQEFKCSGLTEHAPMECGAENSCPHPCRCADGIVDCREKSLTSVPVTL
PDDTTDVRLEQNFITELPPKSFSSFRRLRRIDLSNNNISRIAHDALSGLKQLTTLVLYGNKIKDLPSGVFKGLGSLRLLL
LNANEISCIRKDAFRDLHSLSLLSLYDNNIQSLANGTFDAMKSMKTVHLAKNPFICDCNLRWLADYLHKNPIETSGARCE
SPKRMHRRRIESLREEKFKCSWGELRMKLSGECRMDSDCPAMCHCEGTTVDCTGRRLKEIPRDIPLHTTELLLNDNELGR
ISSDGLFGRLPHLVKLELKRNQLTGIEPNAFEGASHIQELQLGENKIKEISNKMFLGLHQLKTLNLYDNQISCVMPGSFE
HLNSLTSLNLASNPFNCNCHLAWFAECVRKKSLNGGAARCGAPSKVRDVQIKDLPHSEFKCSSENSEGCLGDGYCPPSCT
CTGTVVACSRNQLKEIPRGIPAETSELYLESNEIEQIHYERIRHLRSLTRLDLSNNQITILSNYTFANLTKLSTLIISYN
KLQCLQRHALSGLNNLRVVSLHGNRISMLPEGSFEDLKSLTHIALGSNPLYCDCGLKWFSDWIKLDYVEPGIARCAEPEQ
MKDKLILSTPSSSFVCRGRVRNDILAKCNACFEQPCQNQAQCVALPQREYQCLCQPGYHGKHCEFMIDACYGNPCRNNAT
CTVLEEGRFSCQCAPGYTGARCETNIDDCLGEIKCQNNATCIDGVESYKCECQPGFSGEFCDTKIQFCSPEFNPCANGAK
CMDHFTHYSCDCQAGFHGTNCTDNIDDCQNHMCQNGGTCVDGINDYQCRCPDDYTGKYCEGHNMISMMYPQTSPCQNHEC
KHGVCFQPNAQGSDYLCRCHPGYTGKWCEYLTSISFVHNNSFVELEPLRTRPEANVTIVFSSAEQNGILMYDGQDAHLAV
ELFNGRIRVSYDVGNHPVSTMYSFEMVADGKYHAVELLAIKKNFTLRVDRGLARSIINEGSNDYLKLTTPMFLGGLPVDP
AQQAYKNWQIRNLTSFKGCMKEVWINHKLVDFGNAQRQQKITPGCALLEGEQQEEEDDEQDFMDETPHIKEEPVDPCLEN
KCRRGSRCVPNSNARDGYQCKCKHGQRGRYCDQGEGSTEPPTVTAASTCRKEQVREYYTENDCRSRQPLKYAKCVGGCGN
QCCAAKIVRRRKVRMVCSNNRKYIKNLDIVRKCGCTKKCY
Previous | Next | Top


