Frameshifts
The horizontal sequence is a human 3' EST coding
for an unidentified protein (well, it has now been
identified, but let's pretend it had not...). The
vertical is the protein sequence of the closest
BLASTX match, in this case a mouse
granulocyte colony stimulating factor receptor
precursor (whew! :-).
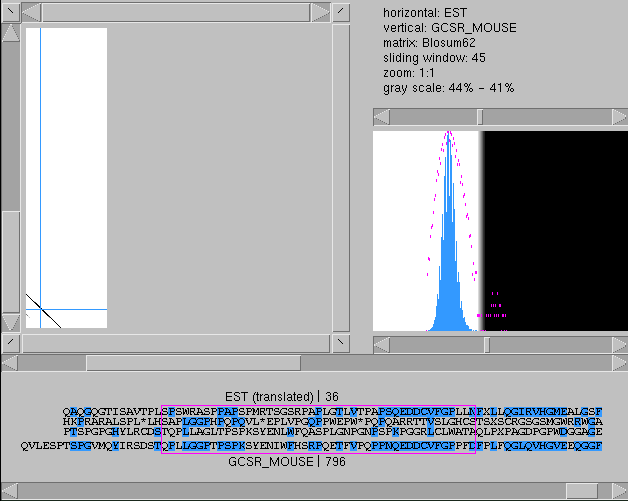
As we can see from the graph, the EST matches
the C-terminal part of the protein and contains
a 3'-UTR. The diagonal is not perfect, however:
there are two partial diagonals but the C-terminal
one is shifted one position downstream with
respect to the first one. This is a clear sign
of a frameshift. The alignment window shows this
well: there is a rather good match on frame 1
but it starts at position 806 or so (in the
protein sequence).
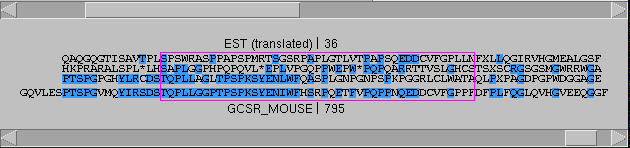
If we move the protein sequence one residue
upstream, we get another good match, but this
time on frame 3, and it extends only through
position 792 (of the protein sequence). It might
seem strange that the match on frame 1 doesn't
start where the match on frame 3 ends, but this
is due to sequence divergence, not to the
frameshift..
- EST:
GCCCCACAAGCCCAGGGCCAGGGCACTATCTCCGCTGTGACTCCACTCAGCCCCTCTTGGCGGGCCTCAC
CCCCAGCCCCAAGTCCTATGAGAACCTCTGGTTCCAGGCCAGCCCCCTTGGGAACCCTGGTAACCCCAGC
CCAAGCCAGGAGGACGACTGTGTCTTTGGGCCACTGCTCAACTTCCCNCTCCTGCAGGGGATCCGGGTCC
ATGGGATGGAGGCGCTGGGGAGCTTCTAGGGCTTCCTGGGGGTTCCCTTCTTGGGCCTGCCTCTTAAAGG
CCTGAGCTAGCTGGGAGAAGAGGGGAGGGTCCATAAAGCCCATTGATTAAAAATTACCCCAGCCCAGGGT
TTTCACCATNTTCCAGTTCACCAGCATCT
- GCSR_MOUSE (P40223):
MVGLGACTLTGVTLIFLLLPRSLESCGHIEISPPVVRLGDPVLASCTISPNCSKLDQQAKILWRLQDEPIQPGDRQHHLP
DGTQESLITLPHLNYTQAFLFCLVPWEDSVQLLDQAELHAGYPPASPSNLSCLMHLTTNSLVCQWEPGPETHLPTSFILK
SFRSRADCQYQGDTIPDCVAKKRQNNCSIPRKNLLLYQYMAIWVQAENMLGSSESPKLCLDPMDVVKLEPPMLQALDIGP
DVVSHQPGCLWLSWKPWKPSEYMEQECELRYQPQLKGANWTLVFHLPSSKDQFELCGLHQAPVYTLQMRCIRSSLPGFWS
PWSPGLQLRPTMKAPTIRLDTWCQKKQLDPGTVSVQLFWKPTPLQEDSGQIQGYLLSWNSPDHQGQDIHLCNTTQLSCIF
LLPSEAQNVTLVAYNKAGTSSPTTVVFLENEGPAVTGLHAMAQDLNTIWVDWEAPSLLPQGYLIEWEMSSPSYNNSYKSW
MIEPNGNITGILLKDNINPFQLYRITVAPLYPGIVGPPVNVYTFAGERAPPHAPALHLKHVGTTWAQLEWVPEAPRLGMI
PLTHYTIFWADAGDHSFSVTLNISLHDFVLKHLEPASLYHVYLMATSRAGSTNSTGLTLRTLDPSDLNIFLGILCLVLLS
TTCVVTWLCCKRRGKTSFWSDVPDPAHSSLSSWLPTIMTEETFQLPSFWDSSVPSITKITELEEDKKPTHWDSESSGNGS
LPALVQAYVLQGDPREISNQSQPPSRTGDQVLYGQVLESPTSPGVMQYIRSDSTQPLLGGPTPSPKSYENIWFHSRPQET
FVPQPPNQEDDCVFGPPFDFPLFQGLQVHGVEEQGGF
Previous | Next | Top


