| Class Name | Symbol | Fields | Description |
|---|
| ActiveSite |  | position, comment | Residue that belongs to an enzyme's active site |
| DNABind |  | begin, end, comment | DNA binding domain |
| Domain |  | begin, end, comment | Generic, unlabeled domain |
| GlycSite |  | position, comment | Glycosylated residue |
| Helix |  | begin, end | Alpha helix |
| LabeledDomain |  | begin, end, comment, label | Generic domain with a user-supplied label. This class has specialized subclasses |
| Lipid |  | position, comment | Lipid-modified residue |
| MetalBind |  | position, comment, symbol | Residue with a metal ligand. symbol is the metal's chemical symbol. |
| ModRes |  | position, comment, type | Covalently modified residue. The type is a single letter denoting the modification type. Valid letters are P (phosphorylation), S (sulfatation), A (acetylation), H (hydroxylation), Y (pyrrolidone carboxylic acid), G (gamma-carboxylation), a (amidation), M (methylation). |
| Mutation |  | position, comment | Point mutation |
| Repeat |  | begin, end, comment | Repeated region |
| SSb |  | begin, end, comment | Intra-chain disulfide bridge |
| Strand |  | begin, end | Beta strand |
| Term | 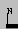 | position, symbol, comment | Terminus. The type is denoted with the one-letter symbol, which can be N or C, but also for example S (start of signal peptide) or P (propeptide), etc. This is also used in DNA sequences for identifying 5' and 3' termini. |
| Transmem |  | begin, end, comment | Transmembrane domain |
| Turn |  | begin, end | Turn (in secondary structure) |
| Variant |  | position, comment | Sequence variant |
| Varsplic |  | begin, end, comment | Splicing variant |
| XSSb |  | begin, end, comment | Inter-chain disulfide bridge |